The University of Manchester provides an environment to integrate basic, clinical and translational research for maximum societal impact.
Our researchers are world-leaders in basic and clinical research who are applying single cell approaches to enhance their research programmes.
This research is supported by state-of-the-art core research facilities containing a range of single cell analysis platforms operated by talented technology specialists.
Why single cell?
- Single cell research is becoming increasingly important to study the cellular heterogeneity found in biological systems.
- Single cell-based sequencing approaches are providing insights into the cell types found in healthy and diseased tissues, and during development.
- Cutting-edge high dimensional spatial imaging and transcriptomics approaches are providing new insight into the cellular architecture of tissues and the cellular interactions and networks driving disease.
- These approaches are further augmented by studies at the protein level based on flow cytometry and mass cytometry.
- Advanced microscopy approaches coupled to fluorescent/bioluminescent markers are providing insights into how single cells behave in complex environments.
Featured projects
See a selection of current research projects using single cell approaches to understand disease.
Mapping the biological drivers of fibrosis formation in tissues
Fibrosis (or scarring) is a common step in the majority of chronic diseases occurring in any organ. The mechanisms driving fibrosis are complex involving many different cell types. To capture the genome-wide spatial distribution of transcripts in relation to disease pathology in fibrosis, we have used spatial transcriptomics (ST) and characterized cellular heterogeneity at near single-cell resolution, while preserving spatial information. Through computational approaches, we have integrated single cell RNA and ATAC sequencing datasets to define cell type clusters and deconvolute multi-cell ST data. These studies have revealed spatial molecular signatures of scar-associated and impaired regenerative cell sub-populations associated with the pathophysiology of fibrotic disease.
Researchers
Delineating transcriptional regulation of tissue-resident immune cells
Tissue resident immune cells within organs such as the gastrointestinal tract, lung and skin play critical roles in maintaining mammalian health, or driving chronic disease. The way in which immune cells respond to their environment or infectious challenge is dictated by transcription factors and tissue specific signals that alter the cellular programme at the level of mRNA. Using single cell technologies - such as scRNA seq - we are able to dissect the transcriptional pathways that determine whether an immune cell is inflammatory or protective in a range of tissues and clinical settings. Such advances provide us with unparalleled resolution and insights into the way the immune system functions and allows us to identify novel targets for therapeutic intervention.
Researchers
Understanding how genetic variants increase disease risk
Most common diseases like arthritis or psoriasis have a strong genetic component. Understanding how genetic variation leads to disease can lead to the identification of novel drug targets, risk prediction and personalised medicine. Hundreds of genetic risk variants have been identified for these diseases, but they do not affect protein coding genes directly and, as a result, it is difficult to disentangle the mechanisms by which they cause disease. Our research uses single cell technologies, genetics, multiomics integration and genome editing to understand how non-coding variation alters the regulation of gene expression in tissues affected by disease (blood, joints, skin, among others), with the aim of bridging the gap between genetic discovery and clinical application.
Researcher
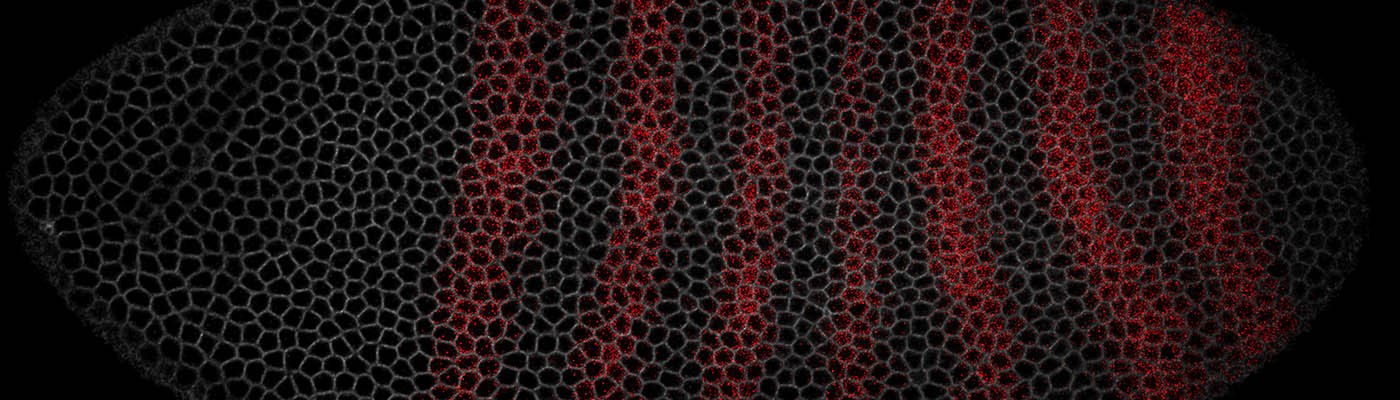
Dynamics of mRNA accumulation and translation in embryonic development at single cell resolution.
The ability of cells to generate complex gene expression patterns is fundamental to multicellular life. While global mRNA levels can be routinely determined using ‘omics technologies, a major gap in our knowledge is how a cell produces these mRNA levels with the correct temporal dynamics. We are exploiting the rapid development and tractability of Drosophila to address how dynamic regulation of different steps in the gene expression pathway drives developmental patterning. We use live imaging approaches to visualize and model spatiotemporal gene expression dynamics at single cell resolution during embryogenesis. The effect of perturbing the dynamics on the accuracy and robustness of embryonic patterning is then determined.
Researchers
A cell atlas of the human outflow tract of the heart.
The outflow tract (OFT), which separates to become the aorta and pulmonary trunk, is rapidly formed and remodelled over a three-week period during human embryogenesis.
Defects arising in this process account for a third of all cases of chronic heart disease, yet little is currently known about the molecular signals orchestrating this critical event.
We are using massively parallel single nucleus RNA sequencing (snRNA-seq) and single cell assay for transposase-accessible chromatin (scATAC-seq) on cells from developing and adult hearts to define the different cell types that form the OFT and the gene regulatory networks that control their regulation.
Researchers
Decoding cell lineages in human oesophagus, stomach and duodenal development.
This study builds on our recent findings that Barrett’s oesophagus, which is associated with progression to oesophageal adenocarcinoma, represents a reversion of normal stratified epithelium to a primitive developmental gut-like state.
We will use single cell transcriptomic and chromatin mapping techniques to compare the gene expression signatures of cells from different developmental timepoints with adult cells, and define the cell types and pathways controlling their differentiation in the upper gastrointestinal tract.
These insights will be crucial in finding a cure for what remains an aggressive and largely incurable cancer.
Researchers
Dynamic cell fate decisions in the developing eye.
In recent years, our understanding of how stem cells make fate transitions has been transformed by the application of single cell technologies.
New single cell live imaging approaches are revealing the importance of protein expression dynamics, particularly pulsatile and short-period oscillatory expression of the Hes genes, on the control of cell fate.
We use single-cell live imaging in combination with mathematical modelling to explore how mechanical signals are integrated with gene expression dynamics in the developing eye.
Researcher
Highlight publications
Browse a selection of our research papers.
Hematopoietic stem and progenitor cells are present in healthy gingiva tissue.
Authors: Siddharth Krishnan, Kelly Wemyss, Ian E Prise, Flora A McClure, Conor O'Boyle, Hayley M Bridgeman, Tovah N Shaw, John R Grainger, Joanne E Konkel
Journal: Journal of Experimental Medicine
Reciprocal transcription factor networks govern tissue-resident ILC3 subset function and identity.
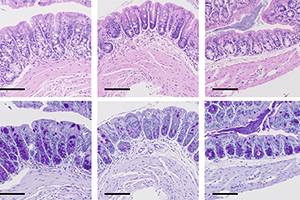
Authors: Rémi Fiancette, Conor M. Finlay, Claire Willis, Sarah L. Bevington, Jake Soley, Sky T. H. Ng, Syed Murtuza Baker, Simon Andrews, Matthew R. Hepworth & David R. Withers
Journal: Nature Immunology
Chronic myelomonocytic leukaemia stem cell transcriptomes anticipate disease morphology and outcome.

Authors: Daniel H Wiseman, Syed M Baker, Arundhati V Dongre, Kristian Gurashi, Joanna A Storer, Tim CP Somervaille, Kiran Batta.
Jounal: EBioMedicine, Volume 58, 2020, 102904.
Quantitative single-cell live imaging links HES5 dynamics with cell-state and fate in murine neurogenesis.

Authors: Cerys S. Manning, Veronica Biga, James Boyd, Jochen Kursawe, Bodvar Ymisson, David G. Spiller, Christopher M. Sanderson, Tobias Galla, Magnus Rattray & Nancy Papalopulu
Journal: Nature Communications, volume 10, Article number: 2835 (2019)
ZIC3 controls the transition from naive to primed pluripotency.

Authors: Shen-Hsi Yang, Munazah Andrabi, Rebecca Biss, Syed Murtuza Baker, Mudassar Iqbal, Andrew D. Sharrocks.
Journal: Cell Reports, Volume 27, Issue 11, 11 June 2019, Pages 3215-3227.e6
Classifying cells with Scasat, a single-cell ATAC-seq analysis tool.

Authors: Syed Murtuza Baker, Connor Rogerson, Andrew Hayes, Andrew D Sharrocks, Magnus Rattray.
Journal: Nucleic Acids Research, Volume 47, Issue 2, 25 January 2019, Page e10
Decoupling the roles of cell shape and mechanical stress in orienting and cueing epithelial mitosis.

Authors: Alexander Nestor-Bergmann, Georgina A. Stooke-Vaughan, Georgina K. Goddard, Tobias Starborg, Oliver E. Jensen, Sarah Woolner.
Journal: Cell Reports, Volume 26, Issue 8, 2019, Pages 2088-2100.e4, ISSN 2211-1247
Quantitative analysis of competitive cytokine signaling predicts tissue thresholds for the propagation of macrophage activation.

Authors: James Bagnall, Christopher Boddington, Hazel England, Ruth Brignall, Polly Downton, Zainab Alsoufi, James Boyd, William Rowe, Alexander Bennett, Catherine Walker, Antony Adamson, Nisha M. X. Patel, Ronan O’Cualain, Lorraine Schmidt, David G. Spiller, Dean A. Jackson, Werner Müller, Mark Muldoon, Michael R. H. White, Pawel Paszek
Journal: Science Signaling, Volume 11, Issue 540, July 2018
Quantitative analysis reveals crosstalk mechanisms of heat shock-induced attenuation of NF-kB signaling at the single cell level.

Authors: Kardyńska M, Paszek A, Śmieja J, Spiller D, Widłak W, White MRH, et al.
Journal: PLoS Comput Biol 14(4): e1006130, 2018
GrandPrix: Scaling up the Bayesian GPLVM for single-cell data.

Authors: Sumon Ahmed, Magnus Rattray, Alexis Boukouvalas
Journal: Bioinformatics, Volume 35, Issue 1, 01 January 2019, Pages 47–54, 2018
Identifying stochastic oscillations in single-cell live imaging time series using Gaussian processes.

Authors: Nick E. Phillips, Cerys Manning, Nancy Papalopulu, Magnus Rattray
Journal: PLOS Computational Biology 13(5): e1005479, 2017
Modulation of promoter occupancy dictates the transcriptional response to graded BMP signalling levels in the Drosophila.

Authors: Caroline Hoppe, Jonathan R. Bowles, Thomas G. Minchington, Catherine Sutcliffe, Priyanka Upadhyai, Magnus Rattray, Hilary L. Ashe
Journal: Developmental Cell, Volume 54, Issue 6, 2020, Pages 727-741.e7, ISSN 1534-5807
Technologies
Our single cell equipment is housed within our world-leading technology platforms.
For single cell sequencing we have two platforms, the 10X Genomics chromium and the Takara ICELL8 system. These feed in to our comprehensive Illumina-based sequencing platforms.
We also have a CyTOF and Imaging CyTOF, and multicolour FACS Symphony, LSRFortessa and spectral Bigfoot flow cytometry machines for for multiplex analysis of protein expression in single cells in cell populations and tissues.
For cellular analysis, we have a suite of microscopes, including 2 Zeiss LSM800 confocal systems with AiryScan detectors, and an LSM780 and LSM710, all capable of advanced fluorescence correlation spectroscopy (FCS) and Raster Image Correlation Spectroscopy (RICS) for single molecule analysis of protein quantification, dynamics and interactions. We also have an upright Leica SP8 two-photon microscope and 2 Zeiss Light Sheet microscopes allowing high-definition in vivo and in vitro investigation of cells within tissue environments.
All of the above are complemented by outstanding support from our team of experienced bioinformaticians, providing data analysis and multi-modal data integration pipelines.
Opportunities at Manchester
We welcome new principal investigators who wish to integrate this fast-moving field into their research area.
The University has invested heavily in single cell biology, and appointed six Presidential Fellows in our first two rounds of recruitment (Matthew Birket, Madhvi Menon, Fong Kuan Wong, Rachel Jennings, Cerys Manning and Andreas Sagner).
We are also committed to training the next generation of scientists in single cell techniques, particularly through opportunities with our BBSRC and MRC DTP, and Wellcome Immunomatrix in Complex Disease PhD programmes.
Our excellence in single cell research has been recognised by the University and has been awarded funds to work towards Institute status. This has been further bolstered by strategic funding from the Wellcome Trust Institutional Support Fund to build and expand our single cell research base.
Hub members
Find out who is working on single cell research at The University of Manchester.